我使用GA包来最小化一个函数。下面是我实施的几个阶段。
0.库和数据集
library(clusterSim) ## for index.DB()
library(GA) ## for ga()
data("data_ratio")
dataset2 <- data_ratio
set.seed(555)
1.二进制编码并生成初始填充。
initial_population <- function(object) {
## generate a population where for each individual, there will be number of 1's fixed between three to six
population <- t(replicate(object@popSize, {i <- sample(3:6, 1); sample(c(rep(1, i), rep(0, object@nBits - i)))}))
return(population)
}
2.适应度函数使Davies Bouldin(DB)指数最小化。
DBI2 <- function(x) {
## number of 1's will represent the initial selected centroids and hence the number of clusters
cl <- kmeans(dataset2, dataset2[x == 1, ])
dbi <- index.DB(dataset2, cl=cl$cluster, centrotypes = "centroids")
score <- -dbi$DB
return(score)
}
3.用户定义的交叉运算符。这种交叉的方法将避免没有集群被“打开”的情况。伪代码可以在这里找到。
pairwise_crossover <- function(object, parents){
fitness <- object@fitness[parents]
parents <- object@population[parents, , drop = FALSE]
n <- ncol(parents)
children <- matrix(as.double(NA), nrow = 2, ncol = n)
fitnessChildren <- rep(NA, 2)
## finding the min no. of 1's between 2 parents
m <- min(sum(parents[1, ] == 1), sum(parents[2, ] == 1))
## generate a random int from range(1,m)
random_int <- sample(1:m, 1)
## randomly select 'random_int' gene positions with 1's in parent[1, ]
random_a <- sample(1:length(parents[1, ]), random_int)
## randomly select 'random_int' gene positions with 1's in parent[1, ]
random_b <- sample(1:length(parents[2, ]), random_int)
## union them
all <- sort(union(random_a, random_b))
## determine the union positions
temp_a <- parents[1, ][all]
temp_b <- parents[2, ][all]
## crossover
parents[1, ][all] <- temp_b
children[1, ] <- parents[1, ]
parents[2, ][all] <- temp_a
children[2, ] <- parents[2, ]
out <- list(children = children, fitness = fitnessChildren)
return(out)
}
4.突变。
k_min <- 2
k_max <- ceiling(sqrt(75))
my_mutation <- function(object, parent){
pop <- parent <- as.vector(object@population[parent, ])
for(i in 1:length(pop)){
if((sum(pop == 1) < k_max) && pop[i] == 0 | (sum(pop == 1) > k_min && pop[i] == 1)) {
pop[i] <- abs(pop[i] - 1)
return(pop)
}
}
}
5.把碎片拼在一起。使用轮盘选择,交叉问题。=0.8,突变问题。=0.1
g2<- ga(type = "binary",
population = initial_population,
fitness = DBI2,
selection = ga_rwSelection,
crossover = pairwise_crossover,
mutation = my_mutation,
pcrossover = 0.8,
pmutation = 0.1,
popSize = 100,
nBits = nrow(dataset2))
我已经创建了我的初始种群,对于种群中的每个个体,将有固定在3到6之间的1的数量。交叉和变异运算符旨在确保解决方案不会有太多的集群(1)被开启。在整合之前,我已经分别尝试了我的交叉和突变功能,它们似乎运行良好。
理想情况下,最终的解决方案将从初始种群中获得1-=1的数量,也就是说,如果一个个体的染色体中有三个1,那么它最终将随机拥有两个、三个或四个1's.但是我得到了这个解决方案,它显示了12个集群(1's)被“开启”,这意味着交叉和突变操作符运行得很好。
> sum(g2@solution==1)
[1] 12
这里的问题可以通过复制所有代码来重现。有熟悉GA套餐的人可以帮我吗?
[编辑]
尝试使用不同的数据集iris,导致我出现以下错误。(仅更改了数据,其余设置保留)
0.库和数据集
library(clusterSim) ## for index.DB()
library(GA) ## for ga()
## removed last column since it is a categorical data
dataset2 <- iris[-5]
set.seed(555)
> Error in kmeans(dataset2, centers = dataset2[x == 1, ]) :
initial centers are not distinct
我试着查看代码,发现这个错误是由if(any(重复的(中心)))引起的。这可能意味着什么?
有几点值得一提:
>
在交叉中,为了随机选择父[1中带有1的random_int基因位置,]将以下代码行从
random\u a
到
random\u a
另一方也是如此。
然而,我认为这种交叉策略可以保证,任何后代最多可以打开集群比特的总数,作为其父母的最大1比特数(在这种情况下,可以是初始种群的6比特,如果你想在解决方案基因中只差1比特,不应该是4吗?)。
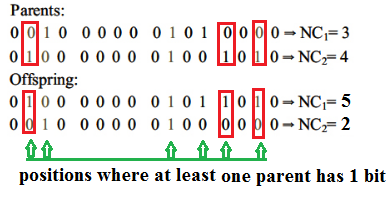
下图显示了3个随机选择的位置,其中至少一个父基因有1位,而交叉和子代产生。

在突变函数中,我认为,更明确地说,我们应该改变这行代码
if((总和(pop==1)
通过
if((总和(pop==1)
用适当的括号。
此外,似乎您的适应度函数(Davies-Bouldin的索引测量集群分离)倾向于打开更多集群。
最后,我认为是突变是罪魁祸首,如果你将k_max更改为一个低值(例如,3)和pmutation更改为一个低值(例如,pmutation=0.01),你会发现在最终的解决方案中,所有基因都开启了4位。
[编辑]
set.seed(1234)
k_min = 2
k_max = 3 #ceiling(sqrt(75))
#5. Putting the pieces together. Using roulette-wheel selection, crossover prob. = 0.8, mutation prob. = 0.1
g2<- ga(type = "binary",
population = initial_population,
fitness = DBI2,
selection = ga_rwSelection,
crossover = pairwise_crossover,
mutation = my_mutation,
pcrossover = 0.8,
pmutation = 0.01,
popSize = 100,
nBits = nrow(dataset2))
g2@solution # there are 6 solution genes
x1 x2 x3 x4 x5 x6 x7 x8 x9 x10 x11 x12 x13 x14 x15 x16 x17 x18 x19 x20 x21 x22 x23 x24 x25 x26 x27 x28 x29 x30 x31 x32 x33 x34 x35 x36 x37
[1,] 0 0 0 0 0 0 1 0 1 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
[2,] 0 0 0 0 0 0 1 0 0 0 1 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
[3,] 0 0 0 0 0 0 1 0 0 0 1 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
[4,] 0 0 0 0 0 0 1 1 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
[5,] 0 0 0 1 0 0 0 0 0 0 1 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
[6,] 0 0 0 1 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
x38 x39 x40 x41 x42 x43 x44 x45 x46 x47 x48 x49 x50 x51 x52 x53 x54 x55 x56 x57 x58 x59 x60 x61 x62 x63 x64 x65 x66 x67 x68 x69 x70 x71 x72
[1,] 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
[2,] 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
[3,] 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
[4,] 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
[5,] 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
[6,] 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
x73 x74 x75
[1,] 0 0 0
[2,] 0 0 0
[3,] 0 0 0
[4,] 0 0 0
[5,] 0 0 0
[6,] 0 0 0
rowSums(g2@solution) # all of them have 4 bits on
#[1] 4 4 4 4 4 4
[编辑2]